Antibiotic Resistance

What is Antibiotic Resistance?
Antibiotics are chemical substances that kill or suppress the growth of microorganisms. Generally, antibiotics function in one of the five following ways:
- Inhibition of nucleic acid synthesis (e.g. Rifampicin; Chloroquine)
- Inhibition of protein synthesis (e.g. Tetracyclines; Chloramphenicol)
- Action on cell membrane (e.g. Polyenes; Polymyxin)
- Interference with enzyme system (e.g. Sulphamethoxazole)
- Action on cell wall (e.g. Penicillin; Vancomycin)
A bacterium is said to be resistant to an antibiotic if it has the ability to withstand antibiotic treatment. Some bacteria are naturally resistant to specific antibiotics. In other cases, bacteria develop resistance over time to an antibiotic to which they were previously susceptible (Yim, 2007).
History of Antibiotics
Although antibiotic compounds were not isolated until around the 1930’s, the Egyptians 3000 years ago used molds to treat some infections and rashes. As well, the Chinese and various Indian tribes had also used molds to treat infections (http://utut.essortment.com/whatisthehist_rgic.htm).
In the 1890s, two German Scientists, Rudolf Emmerich and Oscar Low, began experimenting with various compounds in an attempt to kill microbes. However, rather than using chemical compounds, they used one type of microbe to kill other infectious bacteria. They marketed the microbe Bacillus pycyoneus, which was isolated from the puss of infected bandages, under the trade name pyocyanase, and although it was successful in killing cholera and other diseases in some patients, it caused their conditions to worsen in others. Although no longer used today, pyocyanase was the first antibiotic drug to be used in hospitals.
The first modern day antibiotic compound was isolated in 1928 by Alexander Fleming. Although, the ability of the mold Penicillium notatum to kill bacteria had been noticed before in the 1890’s, it was not until 1928 that the significance of this discovery was realized when the mold was rediscovered by Alexander Fleming.
Figure 1. Alexander Fleming (http://www.wienerzeitung.at/bilder/dossier/flemming.jpg)
Fleming, who had been trained as a surgeon, had been searching for chemical compounds that could kill infections but would not harm the patient. One day while cleaning up his lab, he noticed that one of his Petri plates, on which he was growing the bacterium Staphylococcus aureus, had become infected with mold. However, not only was the plate infected with mold but wherever the mold was present there was an area around it that no bacteria where growing in. The mold which had infected the plate was known as Penicillium notatum. Although he was able to prove that the mold was able to kill bacteria even with significant dilution, he was unable to isolate the penicillin successfully from the mold. It wasn’t until 1939 that 2 scientists, Ernst Chain and Howard Florey, developed a way to isolate penicillin and used it to treat bacterial infections during the Second World War. For their work on discovering and isolating the first antibiotic, Fleming, Chain, and Florey, were awarded the Nobel prize for Medicine in 1945 (http://library.thinkquest.org/25462/history.html).
Around the same time as Fleming, another doctor, Gerhard Domagk, was working on the creation of a synthetic molecule with antibiotic properties named protonsil. Protonsil was the first in a series of drugs now known today as sulfonamides. These synthetic drugs have one major characteristic in common; they all contain a sulphonamide. Although generally not as effective as the naturally occurring antibiotics, sulfonamides have been used to successfully treat many types of infections such as urinary tract infections (http://utut.essortment.com/whatisthehist_rgic.htm).

Figure 2. sulfonamide Group (http://en.wikipedia.org/wiki/Image:Sulfonamide-group.png)
Mechanisms of Resistance
Mechanisms
In general, bacteria have developed several ways to counteract the introduction of foreign antibiotics, which are encoded by resistance plasmids (see section below) or at a chromosomal level (Madigan and Martinko, 2006):
1. The bacteria do not have the proper cellular structure the drug targets. One needs to be sure on what type of bacteria you are treating, as if you are using a drug that is only functional against a structure only found in Gram-negative bacteria, it would most likely be useless against Gram-positive bacteria
2. The antibiotic may not be permeable through the bacterial membrane. Whether it is the size and/or structure of the antibiotic or whether it’s because of the membrane features of the bacteria, the drug cannot be actively transported into the cell. For example, vancomycin, an antibiotic that acts by inhibiting bacterial cell wall formation and used for the treatment of a wide variety of Gram-positive bacteria, cannot be effectively used against Gram-negative bacteria as certain factors associated with the outer membrane (such as porins, lipopolysaccharides, and vancomycin large size) prevent vancomycin from entering (Caillet and Alix, 1996)
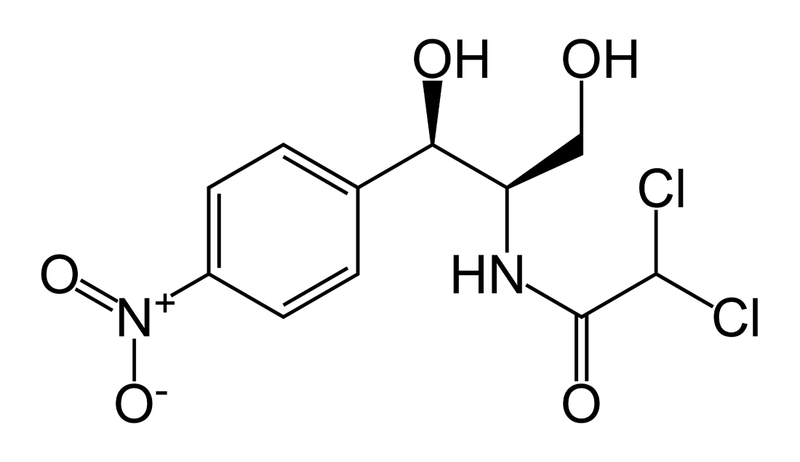
3. The antibiotic may become inactive by modification. There are thousands of enzymes at the disposal of any given bacteria that can simply cut a certain chemical bond, phosphorylate a hydroxyl group, or some other chemical processing that will disrupt the antibiotic and render it inactive. An example of this is the use of chloramphenicol, an antibiotic that acts by preventing peptide bond formation in bacteria, is deactivated when its hydroxyl groups are acetylated by an acetylating enzyme (chloramphenicol acetyltransferase) (Okamoto et al, 1967)
Figure 3. chloramphenicol (photos: http://en.wikipedia.org/wiki/Chloramphenicol)
4. The target of the antibiotic is modified. This will render the antibiotic useless as it will not be able to recognize its active site. An example of this is the use of aminoglycosides, a spectrum of antibiotics that all function by inhibiting the 30S subunit of bacterial ribosomes. Bacteria can develop resistance by a simple mutation to the site where the aminoglycosides bind, preventing aminoglycosides from recognizing their binding site. (http://www.uphs.upenn.edu/bugdrug/antibiotic_manual/aminoglycosideresistance.htm)
5. The bacteria may alter a biochemical pathway in a way which develops resistance to an antibiotic. This means that the bacteria can find alternative routes to making cellular structures, molecules or other biochemical products that are specifically targeted by an antibiotic. An example of this is the resistance of sulfonamides. Sulfonamides function by inhibiting folic acid synthesis in bacteria, which is needed in all cells for growth. Bacteria can move around this problem by taking up folic acid from its environment directly, abandoning the need to synthesize its own folic acid (Madigan and Martinko, 2006)
6. The bacteria have mechanisms to actively displace the antibiotic out of the cell (efflux). Many cases of resistance share this mechanism, as it proves to be quite effective. This can involve anything from specialized proteins that actively transport a drug across the cell memebrane, to proton pumps located on the cell membrane that actively pump out the antibiotic. One of the many examples of this resistance is in tertacyclines, where specialized proton pumps called Tet proteins actively transport tetracycline out of the cell, while bringing H+ protons in (Poole, 2005).
Figure 4. Diagram of Tet protein functionality
Resistance Plasmids
There are many different ways in which a bacterium is able to obtain antibiotic resistance. One of the most common ways is through the acquisition of resistance plasmids. Resistance plasmids are small pieces of DNA that are not part of the bacterial genome; they can either be present in a linear or a circular form. A bacteria can obtain a plasmid through either horizontal or vertical gene transfer. In vertical gene transfer the plasmid is transfered to the offspring during division. However, horizontal gene transfer involves the transfer of genes from one bacterium to another bacterium that is not its offspring. As shown in figure 5 there are 3 main ways that a bacteria can obtain a resistance plasmid through horizontal gene transfer; transformation, conjugation, and transduction (Yim, 2007).

Figure 5. Diagram of the 3 main methods of Horizotal Gene Transfer (Yim, 2007)
Certain types of bacteria are able to undergo natural transformation, which means that they are capable of obtaining small pieces of free DNA from the environment and incorporating it into either their genome or the cell. The DNA obtained is not necessarily a plasmid but if another bacterium containing a resistance plasmid has lysed and that plasmid is not left in the environment it is possible for another bacteria to uptake that plasmid and incorporate it, hence that bacterium which was not previously resistant to a particular antibiotic is now resistant. The next method of horizontal gene transfer is known as conjugation. Conjugation occurs when there is direct cell to cell contact between any two bacteria and a small piece of DNA, usually a plasmid, is transferred from one bacterium to the other. In conjugation the bacteria need not be related, or even the same strain, the only requirement is that both bacteria need to be able to undergo conjugation. The final method of horizontal gene transfer is that of Transduction. Transduction occurs when viruses specific to bacteria, known as bacteriophage, obtain host DNA and transport this DNA to the next bacteria it infects. Usually this only occurs between bacteria which are related (Yim, 2007).
Once the plasmid has been incorporated into the bacterial cell, the plasmid codes for resistance a certain antibiotic according to the mechanisms previously described above. Of the mechanisms listed, as seen in figure 6, there are 3 common mechanisms usually used; antibiotic degrading enzymes, antibiotic altering enzymes, and efflux pumps (Yim, 2007).

Figure 6. The common mechanisms of resistance encoded for by resistance plasmids (Yim, 2007).
Mutagenesis
Originally it was hypothesized that all cases of antibiotic resistance originated from random mutations in the DNA. When a bacterium is ready to divide it replicates its DNA. Although replication of DNA is fairly accurate, a spontaneous mutation can occur with a frequency of 1×10^-7. However, a single mutation does not usually cause the occurrence of antibiotic resistance, therefore usually multiple mutations must occur in order for antibiotic resistance to arise (Yim, 2007).
Recently, research has shown that when bacteria are treated with certain kinds of antibiotics they appear to develop a resistance to these agents faster than by what random mutations can explain (Criz et al, 2005). Two of these antibitics are Ciprofloxacin and Rifampicin.
Ciprofloxacin, or 1-cyclopropyl-6-fluoro-4-oxo-7-piperazin-1-yl-quinoline-3-carboxylic acid, functions by blocking the enzyme DNA gyrase. DNA gyrase or topoisomerase II, is responsible for the supercoiling and uncoiling of DNA. Without the presence of DNA gyrase, this causes double-stranded DNA breaks and no replication of the DNA is able to occur. With the formation of double-stranded DNA breaks, single-stranded DNA is formed. Ciprofloxacin is part of the large group of quinolone antibiotics which are thought to be some of the most important antibiotics available today. Ciprofloxacin was used specifically to treat anthrax during the outbreaks experienced several years ago (Brian, 2001).
Ciprofloxacin
Figure 7. Ciprofloxacin (http://en.wikipedia.org/wiki/Image:Ciprofloxazin.svg)

Figure 8. Rifampicin (http://en.wikipedia.org/wiki/Image:Rifampicin.png)
Because Ciprofloxacin and Rifampicin lead to the formation of single-stranded (ss) DNA, this causes the bacterium to activate a repair mechanism known as the SOS response (Criz et al, 2005). The SOS response is activated when there is an accumulation of ss-DNA at the replication forks where RNA polymerase is unable to bind and protect the ss-DNA. The SOS reponse is a post replication DNA repair mechanism that allows DNA replication to skip over errors and damage that have occurred.

Figure 9. SOS Response ( http://www.science.siu.edu/microbiology/micr460/PageMill%20Images/image36.gif)
Usually the SOS reponse is activated due to damage occurred when the bacteria is exposed to UV light. As seen in figure 9, the UV light causes the formation of dimers in the DNA and this causes RecA to be activated and to cleave the lexA protein. Once lexA protein is cleaved, it activates a series of enzymes which allow replication to occur without the correction of the errors. Hence, this causes an increase in the mutation rate. Although the activation of the SOS-response has mostly been studied in correlation with exposure to UV light, it has also been shown to be activated whenever there is an accumualation of ss-DNA in the cell, as RecA has a high affinity for ss-DNA.
Based on the reasoning that the accumulation of ss-DNA would cause an activation of the SOS-reponse and this in turn, would lead to an increase in the mutation rate. Therefore Cirz et al (2005), set out to see if the increased mutation rate, and hence the increased development of, that occurred upon the addition of Cciprofloxacin and rifampicin was a result of the activation and use of the SOS-response. Using both in vivo and in vitro models, they were able to show that the presence of a functional RNA polymerase and a functional RecA protein is responsible for the rapid rates of antibiotic resistance observed in bacteria treated with these antibiotics (Cirz et al, 2005).
In the in vivo method 6 week old pathogen-free female mice were made immunocompromised through injection with cylcophosphamide. Once their white blood cell count was low enough E. coli broth cultures containing either the wildtype or a mutant in which the SOS-response system has been rendered inactive due to a mutation in LexA, where injected into the thigh muscles of the mice. Following several hours of incubation the mice were then either administered ciprofloxacin or rifampicin. After 24, 48 and 72 hours, 2 mice were sacrificed and their thigh muscles were removed and tested for the presence of non-resistant E. coli and resistant E. coli. As shown in Figure 9 in the LexA mutant the amount of antibiotic resistant bacteria is significantly less than the number of antibiotic resistant bacteria found in the wild type. Therefore, this shows that the SOS-response system is most likely involved in the higher mutation rate, and hence the higher observed antibiotic resistance rate (Criz et al, 2005)
Criz et al, 2005
Figure 10. Adapted from Criz et al, 2005. Survival and Mutation of E. coli Mutants In Vivo after Starting Antibiotic Therapy. Survival and mutation of DlacZ and lexA(S119A) mutants of E. coli ATCC 25922 in thighs of neutropenic (low white blood cell count) mice at 24-h intervals after starting therapy with (A) ciprofoxacin or (B) rifampicin. Open circles and triangles correspond to the total cfu/thigh of the DlacZ and lexA(S119A) strains, respectively. Solid circles and triangles represent the number of drug-resistant DlacZ and lexA(S119A) cfu/thigh, respectively.
The Spread of Resistance
The emergence of antimicrobial resistance in our society is becoming a serious problem. In North America, bacteria strains like Staphylococcus aureus have shown in the past decade to become resistant to antibiotics that are usually prescribed to treat its infections, such as vancomycin and methicillin. We already know that bacteria can be naturally resistant to antibiotics through various mechanisms, R plasmids, mutagenesis, etc., but resistance can be induced by humans, and their improper use of antibiotics. This can lead to widespread resistance to any drug in several bacterial species and the development of super bugs.
One way this occurs is the improper prescription of antibiotics to a patient that a viral infection, which may be due to misdiagnosis from the doctor. Statistics show that approximately only 20% of reported clinical infectious diseases have shown that treatment with antibiotics was necessary and 50% of diagonoses do not prescribe the correct amount of drug (Madigan and Martinko, 2006). The antibiotics prescribed may or may not affect, but it will most likely lead to a few bacteria in the population developing resistance to the drug. We can also say that the misuse of the drug by the patient leads to resistance. For example, if a patient is taking antibiotics for strep throat (colonization by Streptococcus pyogenes), the patient may stop short of using the prescribed amount of drug when he feels better. This will lead to a small percentage of surviving bacteria at the site of infection that have developed resistance to the drug, hence making it harder to treat when it infects the patient again. Another problem is the rampant overuse of one antibiotic in a population. Tons of common antibiotics such as tetracycline, streptomycin and kanamycin have been used worldwide since their discovery, and as such, the amounts of bacterial species resistant to those drugs have increased proportionally (Madigan and Martinko, 2006). Examples of this include the use of penicillin for Neisseria gonorrhoeae, responsible for the STD gonorrhea. Prior to 1980, penicillin was the one of the leading treatments for gonorrhea, which led to the rampant use of the drug. Today, almost all clinical isolations of gonorrhea contain beta-lactamase, the protein that cuts beta-lactam rings and is associated with penicillin resistance. This led doctors to halt the treatment of the STD with penicillin (Madigan and Martinko, 2006). More drugs have been developed to combat gonorrhea, but medicine must come up with new drugs each year, as more and more drugs are becoming resistant.
Figure 11. (from left to right) Streptococcus pyogenes (http://en.wikipedia.org/wiki/Image:Streptococcus_pyogenes_01.jpg), Neisseria gonorrhoeae (http://www.nature.com/emboj/journal/v21/n4/coverfig.gif)
This misuse of antibiotics is not exclusive to humans. Bacterial infections can be easily transmitted though animal to humans in many of our food products that we eat; the most common are Escherichia coli and Salmonella colonization of tainted meat products. For livestock, it is common to use certain antibiotic for growth promoting purposes and for the prevention of disease, not treatment. This has been common clinical veterinary practice for a long time, and due to the tones of antibiotics that have been used in animal feeds, pathogenic bacteria have become resistant to many of these antibiotics, and thus become hard to treat if and when it infects humans. An example of this is the use of fluoroquinones in poultry livestock. Fluoroquinones have been used extensively to prevent respiratory diseases in poultry (Madigan and Martinko, 2006). As a unforeseen side effect of this Campylobacter jejuni, one of the most common causes of food-borne pathogens, has become resistant to the drug (as of 2002 in the US, about 13.5% of all chicken carcasses contain the resistant strain (Lathers, 2002))
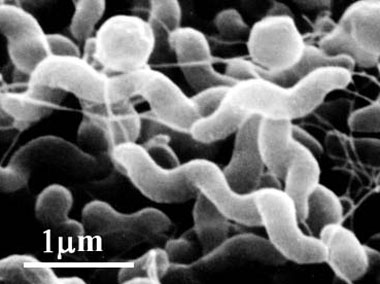
Figure 12. Campylobacter jejuni (http://www.pref.aichi.jp/eiseiken/67f/campylo.jpg)
There is also work currently underway to develop novel drugs that can bock or inhibit some of the mechanisms of resistance. Some of these drugs by themselves don’t actively work to kill bacterial cells, but prevent the mechanics of resistance from functioning properly; therefore potentiating antibiotics that it is taken with. One kind of drug that is being developed is efflux inhibitors. It is believed that the inhibitor will directly block the efflux pump by binding competitively or non-competitively with the pump, but the drug could also act by preventing ATP-binding for ATP-dependent pumps or disrupt the protein gradient of the cell membrane (Marquez, 2005). For example, some tetracycline efflux proteins (namely TetB) have shown to be inhibited through rational drug design by various analogs of tetracycline, such as doxycycline and 13-cyclopentylthio-tetracycline (see photos). Both of these drugs have been proven to be effective against E. coli that have developed resistance to tetracycline, but these drugs are far from universal for treating tetracycline resistance, as there are many structural variations between the various Tet efflux proteins (Marquez, 2005).
Figure 13. (Top) Tetracycline (http://en.wikipedia.org/wiki/Image:Tetracycline_structure.svg), (Botton left to right) TetB efflux protein inhibitors (Marquez, 2005)
Presently, there is a major effort worldwide to stop the widespread resistance that is occurring. Many practical steps for hospitals and medical centers have been laid out by the Centers for Disease Control and Prevention (CDC), which consists of four areas (http://www.cdc.gov/drugresistance/healthcare/ha/12steps_HA.htm):
-Preventing the Infection, by means of vaccinations or minimize use of catheters (medical tubes, IVs, etc.)
–Diagnose and Treat Infection Effectively, by means of targeting the antibiotic specifically and consulting with microbiological experts (i.e. you want to make sure you know what kind of bacterial infection your dealing with, if any)
–Use Antimicrobials Wisely, by making sure your antibiotic treatments are up to date, by using antibiotics susceptibility profiles, by making sure that your treatments are aseptic (i.e., prevent contamination of patient), by making sure that you are treating the specific pathogen and not other colonizing bacteria that may be present in the infection and to know when to stop using the antibiotic
–Prevent transmission, by simply keeping areas around the infection free of contamination and to use basic aseptic techniques such as avoiding contact with people if you are infected and by hand washing.
Combatting Multi-drug Resistance Bacteria
Multi-drug Resistance Tuberculosis and Extensively Drug Resistant Tuberculosis
Tuberculosis (TB) is defined as the colonization of the respiratory tract of the gram-positive, acid-fast bacillus Mycobacterium tuberculosis. This means that the pathogen can be made airborne and can be transmitted by simply allowing the infected to talk. It is one of the leading causes of mortality in our world today, affecting nearly one third of the world’s population (http://www.aafp.org/afp/20051201/2225.html). The pathogen acts in various complex ways with the cells in the host’s lungs in where the initial infection, the bacteria survive and grow in the macrophages located in the lungs (in acute infection cases, there is no host resistance to TB, and rapid lung tissue degradation can occur, which leads to death) (Madigan and Martinko, 2006).
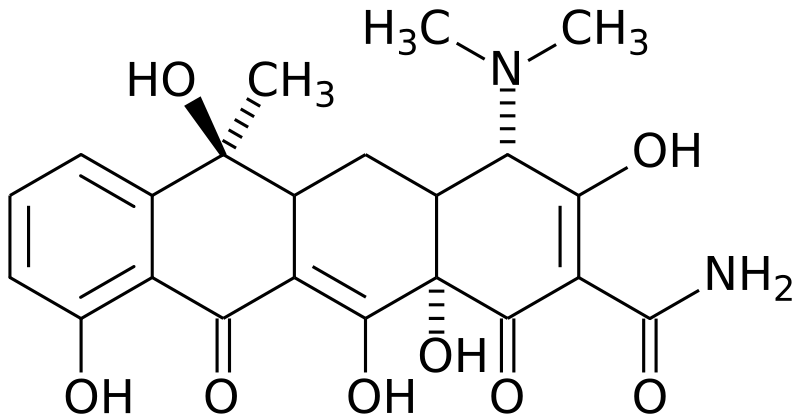
Treatment of a TB infection usually is involved with a combination of first-line drugs, which includes(http://www.aafp.org/afp/20051201/2225.html):
Isoniazid– A prodrug activated by bacterial catalase and affects the synthesis of mycolic acid in TB, which is necessary for cell wall development
Rifampin– inhibits DNA-dependent RNA polymerase which prevent transcription of mRNA
Streptomycin– The first drug discovered to be effective in the treatment of TB. Acts by binding to the ribosomal RNA of the bacterial ribosome, thus preventing protein synthesis
Ethambutol– works by disrupting the cell wall synthesis of the bacteria, which increases the cell wall’s permeability.
Pyrazinamide– A prodrug activated by acidic conditions (coverts to pyrazinoic acid) which can then inhibit an enzyme essential for fatty acid synthesis, which would prevent cell growth.
Figure 14. (Top to bottom) Isoniazid (http://en.wikipedia.org/wiki/Image:Isoniazid_skeletal.svg), Rifampin (http://en.wikipedia.org/wiki/Image:Rifampicin.png)
These drugs are taken in combination with each other to make a quite effective cocktail treatment. Over time, however, treatment with these drugs resulted in many problems of spreading bacterial resistance, such as improper treatment regimens by doctors and hospitals, and failure of patients to take all of the prescribed drugs (http://www.who.int/tb/dots/dotsplus/en/). This usually occurs in societies with poor health care systems (such as third world counties) and can quickly lead to TB strains becoming resistant to any or all of the first-line drugs, meaning we get multi-drug resistant tuberculosis (MDR-TB).
MDR-TB is defined as strains of TB that are at least resistant to Isoniazid and Rifampin, the two most effective first-line drugs. Resistance occurs by mutation of specific genes, whose gene products interact with the drugs. For example Isoniazid is activated by a bacterial catalase, but resistance to Isoniazid occurs when a mutation in the gene katG (whose gene product is the bacterial catalase) and causes the bacterial catalase to not recognize Isoniazid in its prodrug form, meaning Isoniazid will never become active. Preventing the development of MDR-TB should be a major concern to the infected patient, as if resistance occurs to Isoniazid or Rifampin in a regular, non-resistant strain of TB, antibiotic treatment with other first-line drugs can last up to a year, meaning the chances of spreading TB become greater. (Ormerod, 2005)
Before a physician can say a patient has MDR-TB, he needs to be sure that the first-line drugs are ineffective at treating the TB strain. One needs to make an assessment of the dosage of a drug required for each patient individually, as the patient may have been a previous user of TB drugs. This leads to several combinations of first-line drugs that can be given to a patient over months or years. If this is done correctly, this will minimize the chance of MDR-TB evolving from a regular strain of TB (Caminero, 2006) If the patient is indeed confirmed to have MDR-TB, there are several over types of secondary drugs that can be used to treat the strain, all of which are not as effective as Isoniazid or Rifampin. These drugs can also be troublesome due to their side effects or cost (to see a few of the many examples, see (Caminero, 2006) or http://en.wikipedia.org/wiki/MDR-TB). What is important to know is that there is no standard regimen to what drugs should be taken if someone is infected with MDR-TB, which is due to factors like patient sensitivity, toxicity, cost, etc. A last resort strategy to rid MDR-TB if the drugs given are failing is lung surgery, which can be debilitating, but necessary to save the patient’s life (and even then, relapse of MDR-TB can occur after the surgery).
Looking for new Antibiotics
The fight to win against antimicrobial resistance may be a futile one, but newer methods of developing drugs aide in fighting resistant bacterial infections:
Analogs of Existing Drugs
The premise is to create new compounds that are similar to drugs already discovered. This can usually entails optimizing characteristics of the drug, such as solubility and affinity, without making any major modification to the drug’s original molecular structure (Madigan and Martinko, 2006). A good example of this is the tetracycline analogs that are used in treating tetracycline resistance. Methods used to create these analogs usually demands automated processes today (See Combinatorial Chemistry)
Computerized Drug Design
One of the largest areas of research in pharmaceutical industry and one of the most promising prospects in finding novel drugs that are not resistant in certain bacteria. It involves designing drug models using molecular modeling programs that allows the computer to calculate various physical properties of a drug molecule (conformation, bond length, etc.) and see how the drug interacts with its modeled binding site. This costs the industry considerably less and has good predictive power, as some drugs have already been discovered using this method. For example, recently a new drug called R207910, has been discovered to be a potent inhibitor of the ATPase belonging to Mycobacterium tuberculosis. R207910 is a diarylquinoline, and using modeling programs they were able to create the ATPase (using homology modeling) and they were able to see how the drug interacts with its active site on the ATPase model (de Jonge et al., 2007) This drug is one of the newly discovered experimental drugs for potential use against MDR-TB
Figure 15. (from left to right) R207910 (http://en.wikipedia.org/wiki/Image:R207910.svg), Mycobacterium tuberculosis ATPase modelled by de Jonge et al. (2007). The circle represents the binding site for R207910
Other Antimicrobial Agents
Aside from using drugs, bacteriophages (viruses that attack specifically bacteria) have been shown to have potential in treating resistant bacteria. Basically, the virus is engineered to target a specific bacterial species, in which it will inject its DNA, replicate it DNA and kill the bacteria through progeny lysis. The use of bacteriophage has an antimicrobial agent is controversial among the scientific community and is not without its own problems of resistance either, as bacteria can mutate to alter its surface receptors, which will not allow the bacteriophage to recognize its target (Madigan and Martinko, 2006).
Bacteriophage as an Alternative to Antibiotics
It is estimated that every year approximately 44 000 Americans and 8000 Canadians die annually from being infected with an antibiotic resistant strain of bacteria. One of the main problems associated with drug molecules is that the bacteria are able to mutate and the drug molecules are not able to mutate (Riedel, 2005).

Figure 16. Bacteriophage on the surface of an antibiotic resistant bacterial cell (Reidel, 2005)
Also between 1998 and 2003 only 9 new antibiotics received FDA approval, and of those only 2 were new methods of action, where as the other 7 were derivatives of other previously available compounds (Sulakvelidze, 2005).
Unlike drug molecules bacteriophage, which are viruses that attack only bacteria, are able to undergo mutations and are able to kill even the most resistant superbug. Bacteriophage have been used to treat Humans since 1921 but with the discovery and utilization of antibiotics, the use of bacteriophage as a treatment was forgotten in western society. However, research was still continued in non-English speaking countries such as Russia. It is for this reason that although phage may be an alternative to antibiotics, the majority of the research that has been conducted to date has not been done in English. It is hypothesized that until there is a greater translation of the bacteriophage research into English there commercial bacteriophage to treat bacteria will not be available (Sulakvelidze, 2005).
Summary
To give a basic summary of some of the aspects shown, please follow this video link: http://www.sumanasinc.com/scienceinfocus/antibiotics/antibiotics_fla.html
References
-Caminero, J.A. (2006) Treatment of multidrug-resistant tuberculosis: evidence and controversies. The international journal of tuberculosis and lung disease : the official journal of the International Union against Tuberculosis and Lung Disease 10(8):829-37.
-Critz, R.T., Chin, J.K., Andes, D.R., de Creacy-Lagard, V., Craig, W.A., Romesberg, R.E. (2004) Inhibition of Mutation and Combating the Evolution of Antibiotic Resistance. PLoS Biology 3: 1074-1033.
-de Jonge, R. M., Koymans, L.H.M., Guillemont, J.E.G.,Andries, K. (2007) A Computational Model of the Inhibition of Mycobacterium tuberculosis ATPase by a New Drug Candidate R207910. PROTEINS: Structure, Function, and Bioinformatics. 67:971–980.
-Joel Caillet, S. R., and Alix, J.-H. (1996) Amplification of a Novel Gene, sanA, Abolishes a Vancomycin-Sensitive Defect in Escherichia coli. Institut de Biologie Physico-Chimique p. 94-102 [PDF]
-Lathers, C. M., (2002) Clinical pharmacology of antimicrobial use in humans and animals. Journal of Clinical Pharmacology. 42: 587-600
-Madigan, M.T. and Martinko, J.M. 2006. Brock biology of microorganisms. Pearson Prentice Hall. Upper Saddle River, NJ
-Okamoto S, Suzuki Y, Mise K, Nakaya R. (1967) Occurrence of chloramphenicol-acetylating enzymes in various gram-negative bacilli. Journal of Bacteriology. 94(5): 1616-22 [PDF]
-Ormerod, L.P. (2005) Multidrug-resistant tuberculosis (MDR-TB): epidemiology, prevention and treatment. British Medical Bulletin 73-74:17-24
-Poole, K. (2005) Efflux-mediated antimicrobial resistance. Journal of Antimicrobial Chemotherapy. 56: 20–51 [PDF]
-Riedel, G.W. (2005) Alternatives: Phage Therapy: Rediscovering a Treatment for Superbug Infections. The Epoch Times. May 31.
-Sulakvelidze, A. (2005) Phage therapy:an attractive option for dealing with antibiotic-resistant bacterial infections. DDT 10: 807-809.
–http://en.epochtimes.com/news/5-5-31/29150.html
–http://en.wikipedia.org/wiki/Image:Sulfonamide-group.png
–http://en.wikipedia.org/wiki/Image:Ciprofloxazin.svg
–http://en.wikipedia.org/wiki/Image:Rifampicin.png
–http://en.wikipedia.org/wiki/MDR-TB
–http://library.thinkquest.org/25462/history.html
–http://utut.essortment.com/whatisthehist_rgic.htm
–http://www.niaid.nih.gov/factsheets/antimicro.htm
–http://www.science.siu.edu/microbiology/micr460/PageMill%20Images/image36.gif
–http://www.uphs.upenn.edu/bugdrug/antibiotic_manual/aminoglycosideresistance.htm
–http://www.wienerzeitung.at/bilder/dossier/flemming.jpg






“ALL FOR DRUGS” CATERS TO EDUCATION GLOBALLY, No commercial exploits are done or advertisements added by me. This is a compilation for educational purposes only. P.S. : The views expressed are my personal and in no-way suggest the views of the professional body or the company that I represent
142,050 total views, 4 views today





















What’s the gap between a Google Plus page as well as
a profile. If you would like your article to get on-page considered
one of 16,500,000, “make money online” is not the best keyword
to use. Other search engines like yahoo sprang up like mushrooms nevertheless it wasn’t some time before Google stood a stranglehold online, setting itself
up via powerful algorithms because the arbiter of website quality and
importance.